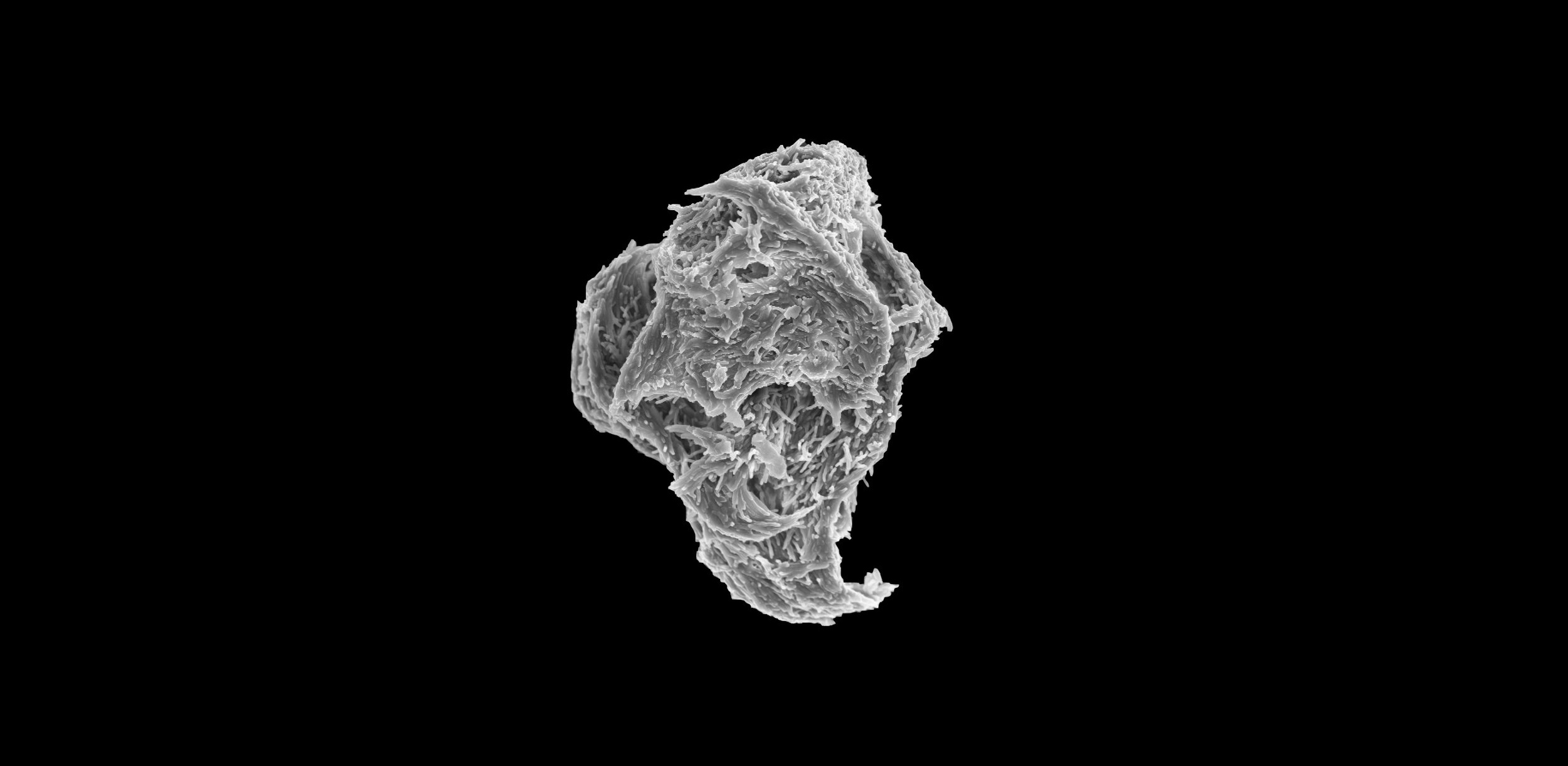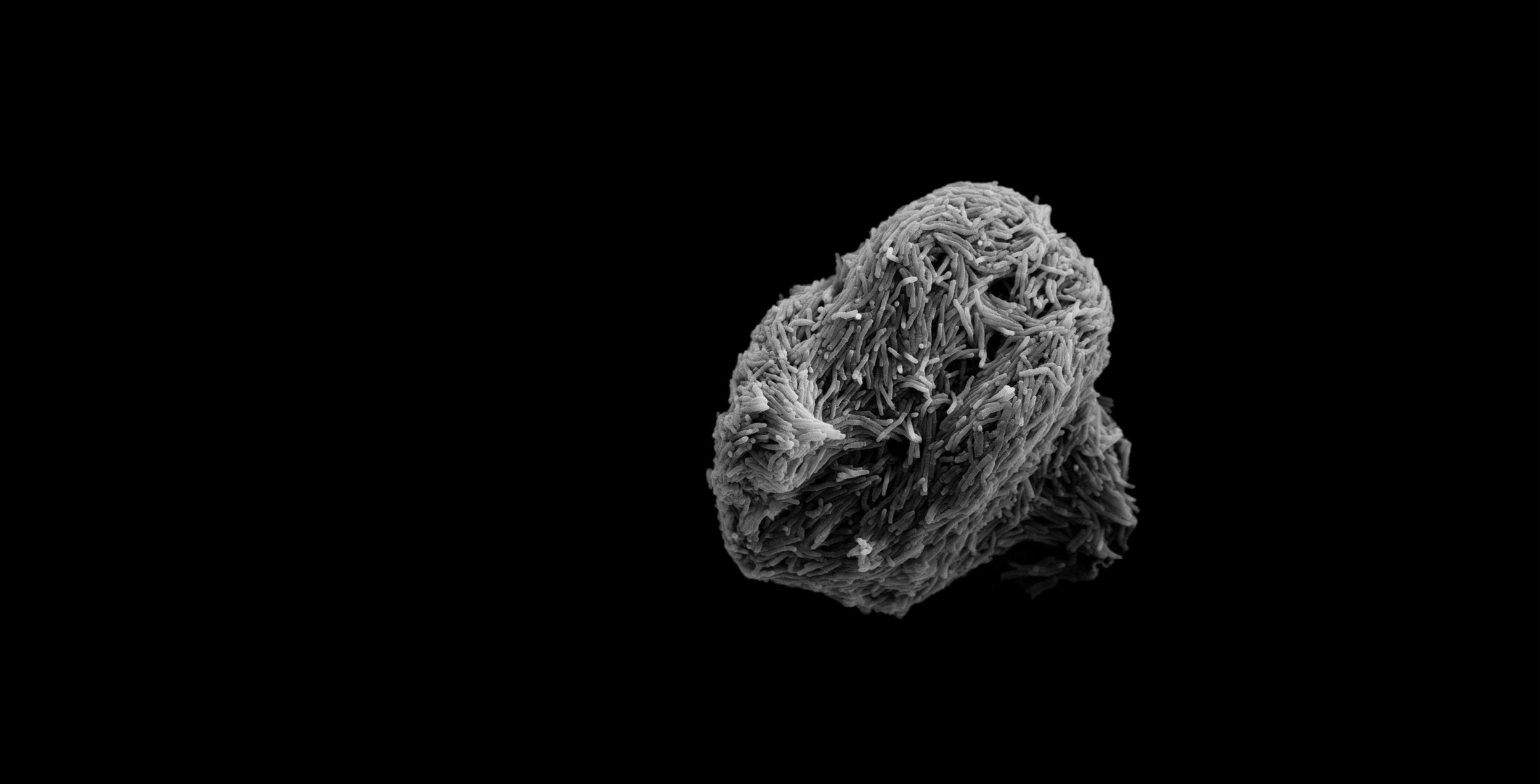
Studying how bacterial communities develop in the host and non-host environments
Research Overview
Bacteria sometimes colonize sites where we don’t want them, most notably in the human body during infection. By thoroughly characterizing the biogeography of these environments while simultaneously developing a mechanistic understanding of species-specific cues that trigger biofilm formation and dispersal, we are working to enhance our ability to control bacterial colonization at any given site.
Nontuberculous Mycobacteria (NTM)
NTM are bacteria that can commonly be found in soils and natural and artificial waterways. They are also emerging opportunistic pathogens that are particularly dangerous to people with underlying lung disorders such as Cystic Fibrosis (CF) or Chronic Obstructive Pulmonary Disease (COPD). The ability of NTM to form multicellular communities helps them survive both in household water systems (a reservoir from which they can infect susceptible hosts) and in animal models of disease. We are therefore working to understand the process by which NTM decides whether to aggregate together or disperse into single cells and whether chemically triggering dispersal would be a viable clinical strategy to counter NTM infections.
Imaging and characterizing microbes in situ
Tissues and other sites of bacterial colonization tend to be opaque, making microscopic analysis over large areas difficult. Our lab utilizes MiPACT-HCR, a combination of tissue-clearing and fluorescent signal amplification techniques, to image bacteria within their native 3D environment. MiPACT-HCR allows us to map interactions, both bacteria-bacteria and bacteria-host, and then to probe cellular activity within the spatial context of the colonization/infection site.
NTM aggregation dynamics in vitro
NTM have a mycolic-acid rich cell wall that has long been thought to confer constitutive aggregation via exposure of a hydrophobic cell surface. Instead, NTM aggregation is regulated at least in part by the ratio of available carbon to available nitrogen. We are interested in the details of this regulation, specifically 1.) What metabolite(s) (intracellular or extracellular) is/are being sensed as the chemical cue for C/N balance? 2.) What surface adhesins are affected by changes in C/N balance? 3.) How is the presence of signal metabolites translated by the cell into changes in surface chemistry? To answer these questions we are employing multiple in vitro biofilm/aggregation assays along with molecular biology techniques.